In plants, metabolic pathways are often compartmentalized within the cell to a greater extent than in the animal kingdom1. This accentuates the importance of proteins that transport metabolites, factors, cofactors or ions across cell boundaries. The transport proteins effectively supply the various anabolic or catabolic pathways of the cell compartments with substrates and because their function may affect the flux of carbon through a metabolic pathway potentially, they represent targets for genetic manipulation2.
Phosphate as a macronutrient
Phosphorus is classified as a macronutrient and an essential element in fertilizers because of its requirement in comparatively large amounts3. However, Pi is the least accessible macronutrient in many ecosystems and its low availability often limits plant growth. Plants have evolved an array of molecular and morphological adaptations to cope with Pi limitation, which include dramatic changes in gene expression and root development to facilitate Pi acquisition and recycling. Although physiological responses to Pi starvation have been increasingly studied and understood, the initial molecular events that monitor and transmit information on external and internal Pi status remain to be elucidated in plants4.
Organic compounds that contain P are used to transfer energy from one reaction to drive another reaction within cells. It is present in every compartment of plant cell and serves a wide range of structural and regulatory roles, including glycolysis modulation, respiration, photosynthesis, starch biosynthesis and sucrose biosynthesis5. Adequate P availability stimulates early plant growth and hastens maturity. In natural systems like soil and water, P exists as phosphate, a chemical form in which each P atom is surrounded by 4 oxygen atoms. However, soil and water (rivers and lakes) usually contain relatively low concentrations of dissolved (or soluble) phosphorus (µM) compared to its concentration inside the plant (mM). P in soil is thought to exist in 3 pools namely solution P, active P and fixed P out of which, it is the solution P pool from which plants take up P in the orthophosphate form (PO4-3). Active P is the solid phase found attached to small particles which are easily converted to solution P pool while fixed pool contains insoluble inorganic phosphate compounds resistant to mineralization by micro organisms in soil3.
Arabidopsis as a model organism

Researchers understood that the study of plants and their development can be made easy with the study of a single plant with similar characters, or rather the need of a model organism. One of the so called model organism is Arabidopsis thaliana, an organism that is easily manipulated, genetically tractable, and about which much is already known. By studying the biology of Arabidopsis, the model plant, we can gain comprehensive knowledge of a complete plant. In the laboratory, Arabidopsis offers the ability to test hypotheses quickly and efficiently6. With the knowledge we gain from the model plant thus established as a reference system, we can move forward with research and rapidly initiate improvements in plants of economic and cultural importance.
Arabidopsis thaliana (Fig.1) is a small flowering plant that belongs to the Brassica family, which includes species like broccoli, cauliflower, cabbage, and raddish. Since Arabidopsis has a small genome relative to other plants and is easily grown under laboratory conditions, it has become the organism of choice for basic studies of the molecular genetics of flowering plants. Scientists expect that systematic studies of Arabidopsis will offer important advantages for basic research in genetics and molecular biology and will illuminate numerous features of plant biology, including those of significant value to agriculture, energy, environment, and human health7.
The commonly used ecotypes are Landsberg erecta, Columbia, and Wassilewskija from NASC8.
Root plastids
Plastids (Fig. 2) are major organelles responsible for photosynthesis, for storage of a variety of products, and for synthesis of key molecules required for the basic architecture and functioning of plant cells9. They have a remarkable ability to differentiate, dedifferentiate and re-differentiate and can be colorless or pigmented at the same time 9 . Proplastids are precursors to all types of plastids from which the starch storing plastids present in roots and storage organs called amyloplasts develop 9 .
The highly studied plastid is the chloroplast and very less on other higher plant non-green plastids because of the fact that considerable expertise is required to prepare intact active non-green plastids free from contaminating membranes and mitochondria10. This kind of preparation is necessary for the accurate investigation of nucleic acids, membrane lipids, protein and enzymes11. Earlier preparations of plastids include aqueous methods involving linear or discontinuous sucrose gradient centrifugation of mechanically prepared crude plastids12 and from broken protoplasts13, 14. But all these methods have problems regarding purity, and intactness and this have been accounted because of the lack of information on root plastid permeability15 even though purification of intact chloroplasts and peroxisomes following centrifugation through a silica-sol pad or gradient has been reported in lots of previous studies16. Later, research came up with methods for isolation and purification of plastids from crude extracts following a low speed centrifugation through Percoll, colloidal silica particles of 15-30 nm diameter coated with polyvinylpyrrolidone (PVP) which renders it non-toxic. Sedimentation occurs at different rates due to its heterogeneity in particle size creating a very smooth isometric gradient17.
Phosphate transporters
Overview
Inorganic phosphate concentration within plant cells is generally less than 10 mM, >3g/kg on a dry weight biomass basis, even though the concentration in typically <10 M18. Because of low concentration of soluble form and slow rate of diffusion to root surface, plants have evolved a range of strategies to increase the availability and uptake of soil Pi18. Research has proved the existence of high and low affinity phosphate transporters responsible for Pi acquisition by plant roots from soil and translocation within the plant respectively18. The high affinity Pi uptake system in plants operates at low Pi concentrations and has an apparent uptake affinity, with Km values ranging from 3-10 µM; whereas the low affinity system operates at high Pi concentrations with Km values ranging from 50-300 µM18.
Studies have revealed the existence of a set of four phosphate translocator (PT) subfamilies with different structures but overlapping specificities, the TPT (triose phosphate/phosphate translocator), GPT (glucose-6-phosphate/phosphate translocator), PPT (phosphoenolpyruvate/phosphate translocator), and XPT (Xylulose-5-phosphate/phosphate translocator)19.
Plastids do not produce PEP even though they are essential for a variety of metabolic pathways confined to plastids like the biosynthesis of amino acids, fatty acids or the precursor for shikimate pathway20, 21. The translocator PPT transports PEP in exchange with Pi released during biochemical processes19. Non-green plastids of heterotrophic tissues like amyloplasts import starch from storage tissues and it relies on the import of hexoseP (Glucose-6-phospahte) 22. This glucose-6-phosphate can act as the precursor of starch biosynthesis or be a part of oxidative pentose phosphate pathway to form trioseP.
Two phosphate uptake systems are known to exist, namely high-affinity and low-affinity uptake systems. The mechanism that depletes Pi with Km values in the µM range active in low Pi concentrations help plants in the uptake of soluble Pi from the soil (no greater than 10µM)23. A high-affinity Pi transporter gene PHO84 was isolated from yeast mutants deficient in Pi absorption3. Later, plant homologs were identified in EST database of the yeast gene and they were used for isolation of cDNA clones from various plant species24, 25, 26. Homologs were also isolated from Arabidopsis thaliana but were designated differently by each group. Eg: PHT1 is identical to AtPT1 and APT2; PHT3 is identical to AtPT4; and PHT4 identical to AtPT2.
The PHT4 family
The PHT family of transporter proteins consists PHT1 (in plasma membrane), PHT2 (plastid inner membrane), PHT3 (mitochondrial inner membrane) 28 and PHT4 (leaves and roots) 29. PHT2; 1, identified in Arabidopsis was found to have similarity with mammalian and fungal Na+/Pi symporters30. But functional analyses in yeast, suggest that PHT2 proteins catalyze H+-dependent Pi transport30, 31, 32. GFP translational fusions with PHT2 proteins in Arabidopsis, Medicago truncatula, spinach and potato have targeted them to the chloroplast envelope, 32, 33, 34 and localization within the inner envelope membrane is affirmed by subcellular proteomics and membrane fractionation / immuno-detection33. In addition to its putative role in Pi import into the chloroplast, the presence of PHT2; 1 transcripts within the root stele suggests that the encoded protein functions in non-photosynthetic plastids34.
The Arabidopsis genome also encodes six PHT4 proteins, all of which mediate Pi transport in yeast with high specificity35. The effects of pH and protonophores on transport activities suggest that PHT4 transporters, like PHT2 proteins, catalyze H+-dependent Pi transport29.
The spatial expression of PHT4 genes have been analyzed with the use of GUS reporter gene where GUS genes are incorporated into their genomes. The transcriptional fusion constructs were introduced into Arabidopsis plants and progeny of the transgenic plants were evaluated for GUS activity using histochemical assays29.
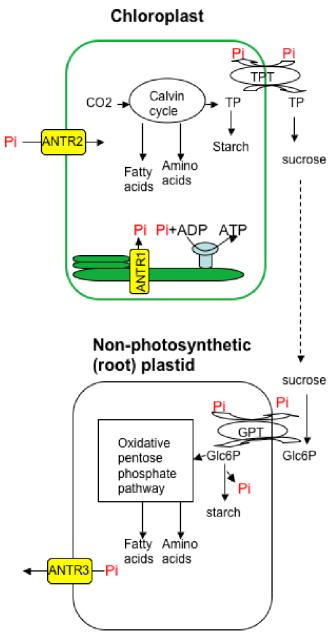
PHT4;1or ANTR1 and PHT4;4 or ANTR2 activity was found throughout green tissues of leaves, seedlings, sepals etc, present in all cell types but only in guard and subsidiary cells of the epidermis. Chlorophyll autofluorescence, co-localized with GUS indicated these proteins to be expressed predominantly in chloroplasts. Even though earlier studies by RT-PCR detected PHT4; 4 in roots35, no GUS activity was detected in roots of these plants. PHT4; 2(ANTR3) was found expressed throughout the roots but not in leaves or other plant parts29. PHT4; 3(ANTR4) and PHT4; 5(ANTR6) expression was restricted to veins of leaves especially in the phloem portion revealed by differential interference contrast (DIC) imaging of transverse leaf sections. PHT4; 3 was detected also at root tips, while PHT4; 6(ANTR5) was localized to the Golgi apparatus29.
Light dependence of these genes was evaluated by monitoring the transcript levels using quantitative RT-PCR and found out that PHT4;1 and PHT4;4 like PHT2;1 transcripts increased rapidly after exposure to light while PHT4;3, PHT4;5 and PHT4;6 had no obvious effects29. PHT4; 2 transcript levels in roots were found to be decreased by about 80 % during the light treatment29. Even though PHT2; 1 expression is not under circadian control, PHT4; 1 exhibits a circadian expression pattern with peak expression during light phase29. Differing from this pattern, PHT4; 4 appeared to be induced by light rather than a circadian rhythm while other members were unaffected29. Transcripts for all these genes were found to be more abundant in light than dark except PHT4; 2 which were rather related to the exposure of the shoot tissues to light29.
Studies indicate that these transporters may serve as two-way valves with transport direction dependent on the Pi electrochemical gradient and proton-motive force29. But in non-photosynthetic plastids where there is no synthesis of ATP, export of Pi not directly coupled to transport of phosphorylated carbon compounds is essential to avoid imbalances29. They have also revealed that PHT4; 2 functions in all root plastids and that Pi transport catalyzed by PHT4; 3 and PHT2; 1 may be needed to supplement that of PHT4; 2 in distinct plastid types, including amyloplasts in root cap columella29.
Calvin cycle takes place in the stroma of chloroplasts; during which carbon dioxide enters the cycle along with the energy provided by photolysis and in a series of steps, glucose is manufactured36. This glucose can act as building blocks in making starch and cellulose or can be used in respiration36. Previous studies have established the presence of phosphate transporters like ANTR1, localized to the thylakoid membrane for export of Pi37 and ANTR2 to the chloroplast inner membrane responsible for import of Pi38. Triose phosphate translocator exports photoassimilates in strict counter-exchange with Pi7. The sucrose produced is transported through phloem, unloaded and broken by invertase or sucrose synthase39. In non-green root plastids, Glucose-6-phosphate translocator imports these molecules which are used for various purposes19. This cycle of events are depicted in our working model (Fig: 3).
Functional Genomics
Bioinformatics makes use of databases to store, organize, generate, retrieve, analyze and share sequences, structures, functions and genetic interactions. Databases include primary databases with primary sequence structure of nucleic acid like EMBL, DDBJ, Gene Bank etc and that of protein like SWISS-PROT, TrEMBL, MIPS, PIR etc. While secondary databases includes secondary structure of proteins like PROSITE, Pfam40.
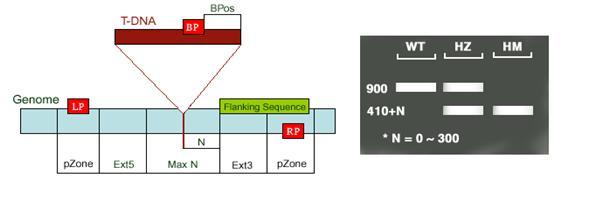
These databases along with other web based tools have provided the first breakthrough regarding the protein ranging from protein sequence to localization, length of targeting peptide, molecular weight, structure, function and sequence homology. Genetic analysis that proceeds from genotype to phenotype using gene manipulation techniques is termed as reverse genetics. Knowledge of the sequence helps in complete inactivation of the gene of interest by designing random mutagens like transposons or T-DNA which can create loss of function in the plants (Fig: 4). Polymerase chain reactions, by designing specific primers for the gene of interest and the insert helps to identify those plants carrying the particular mutation of interest. While screening they are classified as wild type: WT (which do not carry the insert on the gene of interest), homozygous: HM (which has insertions on both the chromosomes) and heterozygous: HT (which carry insertion on just one of the pairs of chromosomes). When PCR is carried out, WT would yield a product corresponding to the size of the gene of interest, HM would yield just one band with a gene specific and T-DNA primer, and HT would yield bands in both using only gene specific and with gene specific and T-DNA primer.
Responsible for this page:
Director of undergraduate studies Biology
Last updated:
05/20/09
