Main Findings of Arabidopsis thaliana nsLTPs Genes
Expression Angler - Bio Array Resource
Figure demonstrates the co expression network of stress up regulated nsLTPs genes, Pearson correlation coefficient (R) was calculated from the microarray analysis of gene expression, the expression angler of the Bio Array Resource, where R=1.00 indicates the bait value and R>0.7 is normally consider as rule-of-thumb threshold for true correlation. Expression Angler collected stress data from the AtGenExpress experiments. The study reveals that nsLTPs form several networks or modules of nsLTP genes showing similar strong co regulation. Hence this indicates that the groups of nsLTP genes involved in the same biological process.

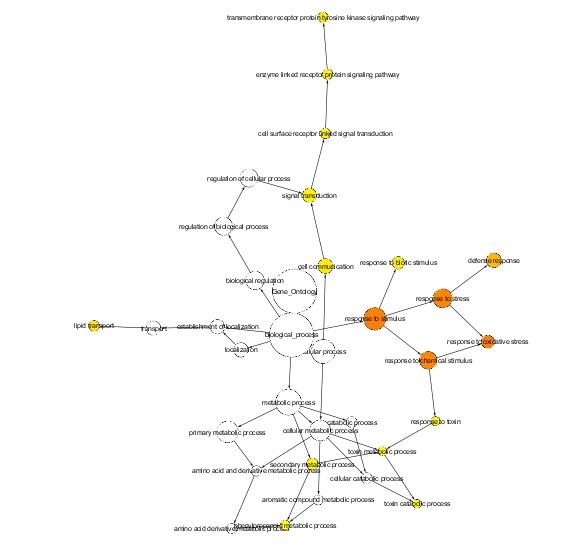
Since genes are expressed in the same module, so we did an enrichment analysis of biological process GO term for each module. We used the same set of 17 genes in the CORNET database. Here the pearson correlation coefficient, R>0.8 was set up and the data was collected from the AtGenExpress experiment. The network was displayed in the graphical format through cytoscape software. Then we implement the BiNGO 2.3 to analyse GO term enrichment, FWER correction was used to control the false positive rate. If the module shows the p value less than 0.05, then the GO term is significantly enriched in the module. Figure shows the significantly over represented GO term in the module II.
Responsible for this page:
Director of undergraduate studies Biology
Last updated:
05/18/10
