Mutants fell into two different categories
Mutants were screened searching for plants with a GFP expression pattern deviating from that found in the AtMC9::nGFP reporter line. When mutants of interest reached ~30 days of age their phenotypes were documented. A mutant with a severely altered growth phenotype would be more likely to carry mutations in genes affecting plant fitness or growth in general without acting directly on expression of metacaspase 9, and were therefore excluded from this study. In the end eleven mutant lineages were selected for further studies.
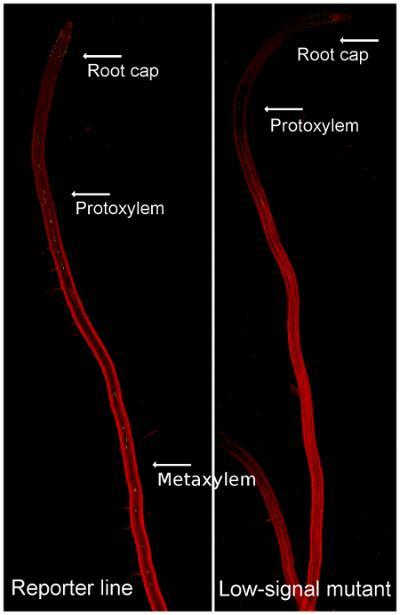
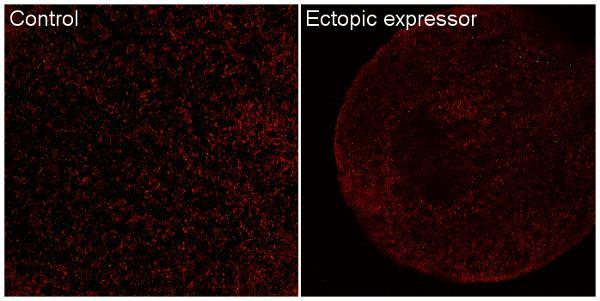
The screening revealed that the mutants could be divided into two different categories: low/no-signal mutants with less GFP in the roots compared to the reporter line and the ectopic expressors with normal GFP signal in the roots and additional signal in the cotyledons. The low-signal mutants could further be divided into low-signal and no-signal mutants depending on how much GFP that could be found in their roots during the initial screen. Confocal microscope showed that the GFP in cotyledons of ectopic expressors was located at the same depth within the tissue as chlorophyll, placing the signal in the mesophyll, and that also the no-signal mutants did have some GFP signal in their roots.
Responsible for this page:
Director of undergraduate studies Biology
Last updated:
05/21/11
