Analysis of allelic diversity of HvNAM-1 gene
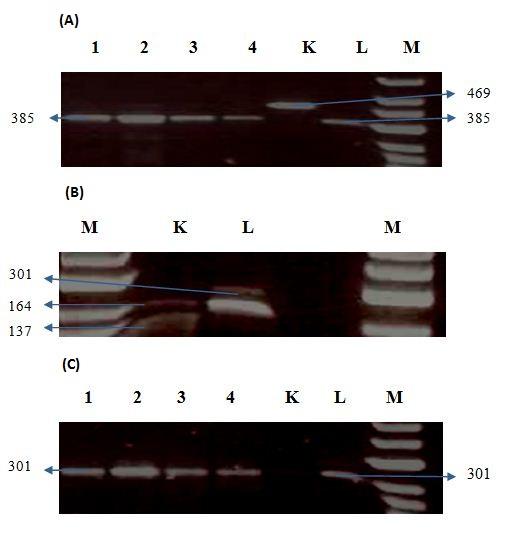
The two CAPS markers uhb6 and uhb7 contain one SNP each between the control cultivars Karl and Lewis. In uhb6 the SNP is located within the third NAC sub-domain of HvNAM-1 and represents the substitution of pro (in Karl) to ala (in Lewis). In uhb7 the SNP is located at the C-terminal end and involves the substitution of thr(in Karl) to ala (in Lewis) HvNAM-1. Based on these combinations, the Karl allele (P in uhb6 and T in uhb7) was named as “PT” and the Lewis allele (A in uhb6 and A in uhb7) was named as “AA” (Distelfeld et al., 2008).
The amplified uhb6 product from Karl produced an uncut 469-bp after digestion with Mwol while a 385-bp fragment was obtained from Lewis (fig.A). Similarly, the digestion of the uhb-7 product (301-bp) with restriction enzyme Taa1 revealed 164+137bp fragments in Karl and a 301bp uncut fragment in Lewis (fig.B). The controls Karl and Lewis were included with the eighty samples to analyse the polymorphism of the allelic diversity of Hv-NAM-1 gene among the four types of cultivars. Our present results show that among the four different types of barley accessions, the Lewis allele “AA”was observed in all the cultivars. The Karl allele “PT” was not observed in any cultivars suggesting lack of polymorphism.
Responsible for this page:
Director of undergraduate studies Biology
Last updated:
05/20/11
