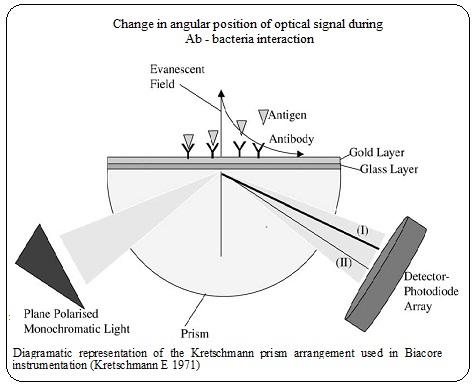
Biofims contain numerous bacteria living in a symbiotic relationship enabling growth conditions suitable for one another and increase the intensity of wounds by impairing healing processes. The identification of pathogens living in biofilms of chronic infections has been difficult with expensive, laborious and time consuming methods like PCR, serological, biochemical and culture techniques. In an attempt to recognize the organisms that are found in biofilms, we have established a SPR-based method to distinguish pathogens by their interaction to specific antibodies. Surface Plasmon Resonance (SPR, Biacore) has been used as a sensing device for label free detection and quantification of macromolecular interactions. Antibodies were immobilised on CM5 sensor chip of Biacore 1000. When there is an interaction between a ligand with it’s specific antigen (bacteria from sample) then there is a change in angular position of the produced optical as a result of change in mass and thus there is a change in refractive index. The interaction could be measured in response units (RU) enabled by Bia-evaluation software. In the present study the results from analysis of ulcer secretion from healthy and patients with chronic ulcer in Biacore was presented. We aimed to develop this method to establish an alternative system for detection of bacteria in poly microbial samples and detect the organisms that might not be discovered by culture or PCR method.
Responsible for this page:
Director of undergraduate studies Biology
Last updated:
05/20/11
