Materials & Methods
Plant material and growth conditions
Arabidopsis seeds were obtained from the Arabidopsis Biological Resource Center (ABRC, Ohio state University, USA). The seeds consisted of Arabidopsis thaliana wild-type (Col-0), AtTPK5-knock-out (SALK_123690_C; tpk5-e) and AtTPK5-knock-down mutant (SALK_113992; tpk5-UTR). The seeds undergo a sterilization process, after which they are placed and grown on 0.7% agar solution in sterilized pipette tip boxes for two weeks. After the initial two weeks all the seedlings are then planted onto frigolite plates under the condition of 120 µmol photons m-2 s-1, 20-25°C, 16 h-dark/8 h-light photo-period with 70% humidity. The nutrition given to the plants contained; 80 ml KNO 3 , 24 ml Ca(NO3)2 , 16 ml KH2PO4 , 16 ml MgSO4 , 16 ml NH4Cl, 16 ml Fe-EDTA, 16 ml µ-solution and 16 ml H20.
Primer design and screening of mutants
The primer design tool at http://signal.salk.edu/tdnaprimers.2.html was used in order to create the primers that would be needed for the screening of homozygous tpk5-e and tpk5-UTR mutants, and the primers were ordered from Invitrogen.
The primers which were used for the screening were the following;
401: 5´-CCTTCTTCCATGAAATCTCCC-3´, 402: 5´- ATTTAACAGCTTTCCGATGCC-3´, 301: 5'-AGCAAGAAAATCGCTTGTCTG-3', 302: 5'-AAGGGATTTGATTTAAAGTTTGTTG-3' and LB 1.3: 5´-ATTTTGCCGATTTCGGAAC-3´
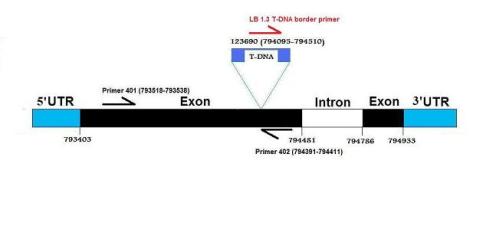
The structure of the AtTPK5 gene and its T-DNA insertion (644 bp) in the first exon of the tpk5-e mutant.
It is also shown where the used primers bind in with their corresponding coordinates.

The structure of the AtTPK5 gene and its T-DNA insertion (644 bp) in the UTR-region of the tpk5-UTR mutant.
It is also shown where the used primers bind in with their corresponding coordinates.
DNA preparation
The sample DNA from the plants was extracted by using a shorty buffer and a TE buffer. The shorty buffer contained: 0.2 M Tris-HCL, 0.4 M LiCl 2 , 25 mM EDTA (pH 8.0), 1% SDS and ddH2O. The contents of the TE buffer were: 10 mM Tris (pH 8.0), 1 mM EDTA (pH 8.0) and ddH2O.
Polymerase Chain reaction (PCR)
The conditions of the PCR program used for the screening of the tpk5-e, tpk5-UTR mutants and wild-type were as follows; 1. Initial denaturation step at 95 º C for 3 min with 1 repeat 2. The second step consisted of denaturation for 30s at 95 º C, annealing with 30s at 52º C and elongation with 60s at 72º C for 30 repeats. 3 A final elongation step was held at 72º C for 10 min. 4. A final holding step at 4 º C
An Oxytherm respirometer was used in assocation with a leaf disc electrode (model LD2, Hansatech, King's Lynn, Norfolk, England) to measure the steady-state photosynthetic oxygen evolution from both the tpk-e mutant and wild-type at 20 °C in a humidified Hansatech electrode chamber (Norfolk, England) which contained a sandwich with different layers consisting of a steel grid, a sponge, steel grid with an unperforated centre, and a steel grid with a perforated centre
Electron Transport rate (ETR)
A Pulse-Amplitude Modulated (PAM)-210 fluorescence measuring device (Walz, Germany) was used to measure the chlorophyll fluorescence and to calculate electron transport rate (ETR) in 4 hour light adapted plants.
Isolation of thylakoids by sucrose graident
Thylakoids were isolated with a sucrose gradient fractionation.
Statistical analysis
All the data were analysed with Student's t-test using open-office, excel and SPSS for the elucidation of differences between the wild-type and mutant.
Responsible for this page:
Director of undergraduate studies Biology
Last updated:
05/21/11
