Introduction
Ecological networks describe the structure of interactions between species in communities and ecosystems. They are used as a tool for analysing ecological communities and comparing them across space and time. One frequently used type of ecological network is the food web, consisting of trophic links between predators and prey. Ecological networks can be used to "model" the effects of interactions between species, and to make predictions for a community's future. For example, networks can be used to make predictions of the effects of invasive species, temperature change and habitat loss.
In each case, the strength of a network-based approach is that it allows researchers to consider all species in the community simultaneously. To do this, however, requires data on all species in the community. These data requirements can make food-web studies difficult as it is sometimes impossible to obtain high-resolution data. Aggregation can be a solution when data is not available for all species. Here I define aggregation as when species are lumped together based on their similarity.
Aggregation can both be a solution and a problem in network analysis. Aggregation can be used to reduce bias in poor-quality data or condensing intractably large datasets. It can also solve problems with spatial and temporal scales.
Despite this, aggregation is known to affect many network properties. One problem caused by aggregation is lumping of specialists and generalists, which makes it difficult to detect the specialists. Another problem is that interactions between species that are lumped together will not be detected. In earlier years, some studies suggested that patterns found might be artifacts of missing species and links in the data due to aggregation.
Previous studies testing effects of aggregation have all focused on network metrics which only describe the network as a whole. These network metrics include connectance (measure the density of links in a network), mean generality (number of prey per predator) and mean vulnerability (number of predators per prey). In this study I analyse the effects of aggregation on the details of network structure. To describe these details, I chose to use motifs.
Aim
Analyse how different degrees of aggregation of network species (nodes) affect network structure, in particular motif frequencies, in a highly resolved food web.
I tested the sensitivity of motif frequencies based on networks generated by aggregating species with similar sets of predators and prey, by aggregating taxonomic groups, and by aggregating subnetworks across space.
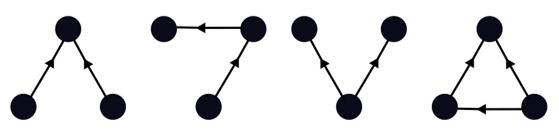
Motifs
Motifs are unique configuration of n interacting species within a network. In this study, I focus on three-species motifs, which give 13 possible motifs. The set of these network motifs can be seen as building blocks of the network, and when reassembled, make up the original network. All motifs correspond to species interactions and some have clear ecological relevance. Examples of motifs that have been studied independently are omnivory, three-species food chain and apparent and direct competition. Motifs can also be used to describe species' roles in networks.
Responsible for this page:
Director of undergraduate studies Biology
Last updated:
05/10/18
